Alternating least squares (ALS) is an algorithm used in recommender systems, which trains the model data X and Y to minimize the cost function as below
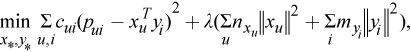
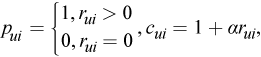
where
- c_ui measures the confidence in observing p_ui
- alpha is the rate of confidence
- r_ui is the element of the matrix R
- labmda is the parameter of the regularization
- n_xu, m_yi denote the number of ratings of user u and item i respectively.
ALS alternatively computes model x and y independently of each other in the following formula:
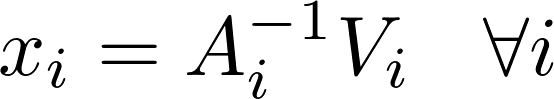
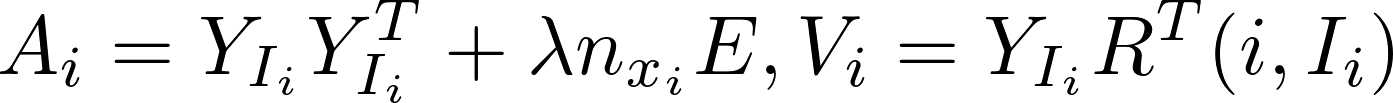
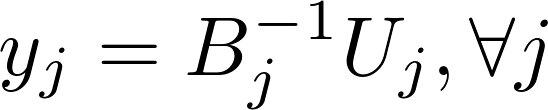
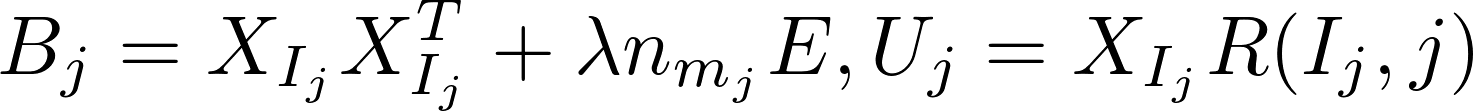
Harp-DAAL currently supports distributed mode of ALS 12 for dense and sparse (CSR format) input datasets.
More algorithmic details from Intel DAAL documentation is here.
- Rudolf Fleischer, Jinhui Xu. Algorithmic Aspects in Information and Management. 4th International conference, AAIM 2008, Shanghai, China, June 23-25, 2008. Proceedings, Springer. [return]
- Yifan Hu, Yehuda Koren, Chris Volinsky. Collaborative Filtering for Implicit Feedback Datasets. ICDM’08. Eighth IEEE International Conference, 2008. [return]